German Congress of Orthopaedics and Traumatology (DKOU 2025)
Deutscher Kongress für Orthopädie und Unfallchirurgie 2025 (DKOU 2025)
Challenges and limitations of large language models in medical information extraction
Text
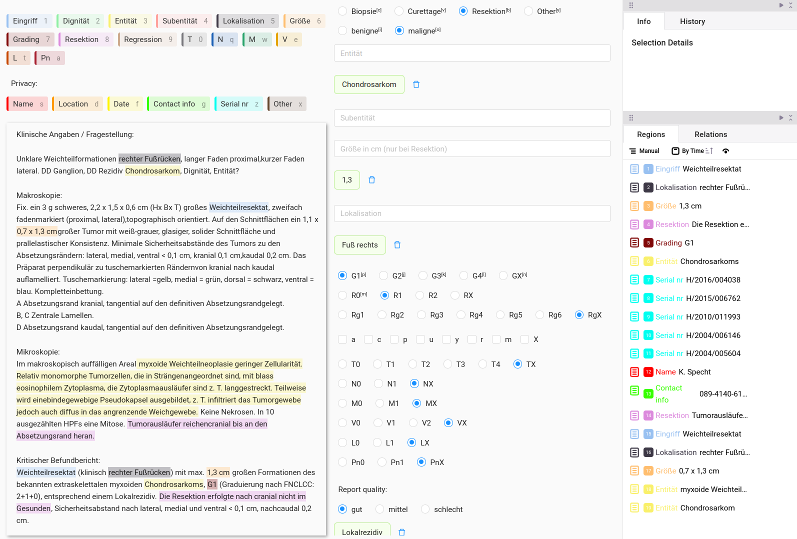
Objectives: Large Language Models (LLMs) have demonstrated remarkable capabilities in general Natural Language Processing (NLP) tasks, sparking interest in their healthcare applications. However, their effectiveness in extracting complex, domain-specific medical information remains uncertain, despite their enormous potential to accelerate clinical workflows (Figure 1 [Fig. 1]). This study evaluates the ability of LLMs to extract key details from clinical records on bone and soft tissue tumors. Our primary research question is: To what extent can LLMs accurately perform medical information extraction tasks without additional training, and how do their limitations impact usability in real-world clinical settings?
Methods: We evaluated the performance of state-of-the-art open-weight LLMs on a retrospective, single-center dataset of annotated clinical reports. These contained essential tumor-related information, including entity, dignity, location, size, grading, and TNM classification. We created a detailed task description and constrained models to produce structured outputs to improve relevance and measurability. We evaluated LLMs in both zero- and few-shot settings, assessing the correctness and appropriateness of the extracted details to determine their reliability for clinical applications.
Results: LLMs demonstrated reasonable performance in extracting general medical information, such as tumor dignity and location, with ~90% F1-score (equal balance of sensitivity and precision, see Table 1 [Tab. 1]). However, this is still inadequate for safe and effective use in clinical settings, especially for domain-specific critical parameters such as tumor grading, resection margins, size, and TNM classification with even lower scores. Combining the detailed task description with example-based prompts further improved performance. These findings indicate that despite the success of LLMs in general NLP tasks, their ability to process intricate medical details remains limited without domain-specific adaptation.
Table 1: Evaluation results for Llama 3.3 across different tumor-related information. 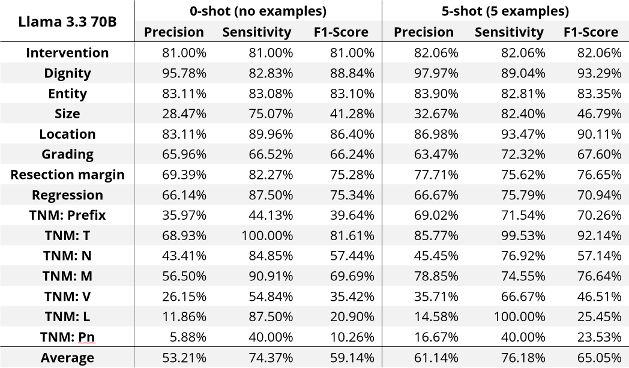
Discussion and conclusions: Our study highlights a fundamental gap between the perceived capabilities of LLMs and their actual performance in medical information extraction. While these models can assist with structured data extraction, their reliability diminishes for nuanced, clinically significant details. Our findings suggest a thorough description of required information integrated with few-shot examples is crucial for enhancing generalization across diverse scenarios. We underscore the necessity of close collaboration between computer scientists and clinicians to define task scopes and structure extraction requirements effectively. Future research should explore lightweight fine-tuning strategies tailored to specific medical subdomains to enhance LLM performance and ensure their practical utility in clinical workflows. For this, high-quality annotations provided by experienced clinicians are indispensable.