- Conferences Overview
- Deutscher Kongress für Orthopädie und Unfallchirurgie 2025 (DKOU 2025)
- Closing the data gap: A fully automated AI pipeline for digitizing orthopedic paper records into structured registries
German Congress of Orthopaedics and Traumatology (DKOU 2025)
28.-31.10.2025
Berlin
Deutscher Kongress für Orthopädie und Unfallchirurgie 2025 (DKOU 2025)
Closing the data gap: A fully automated AI pipeline for digitizing orthopedic paper records into structured registries
Text
Objectives and questions: The digitization of paper-based medical documents presents significant challenges for healthcare institutions. Thousands of unstructured medical documents remain unused in hospital archives, despite containing valuable information for research and treatment optimization. This study aimed to develop an artificial intelligence (AI)-powered pipeline for the automated extraction of clinical variables of interest from scanned medical documents using large language models (LLMs).
Material and methods: A multi-stage processing pipeline was developed, combining optical character recognition (OCR) and LLMs (Figure 1 [Abb. 1]). The pipeline consists of (i) OCR-based text extraction, (ii) LLM-assisted text correction, (iii) LLM-powered extraction of clinically relevant variables, and (iv) storage in a structured database. Various open-source, and proprietary LLMs were evaluated and compared, including OpenAI’s popular ‘GPT-4o’ and ‘GPT-3.5’, as well as Meta’s ‘Llama 3 8B’ and ‘Llama 3 70B’.
Figure 1: Comprehensive pipeline for fully automated digitization of medical documents using optical character recognition and large language models, including the extraction of clinically relevant variables and storage in a structured database format, enabling queries for downstream applications.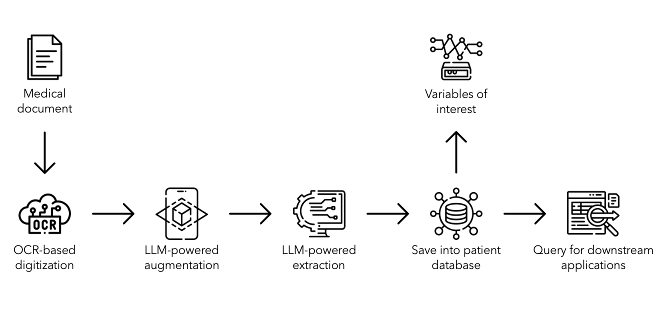
Results: The developed pipeline achieved a near-perfect OCR accuracy of 97.29 ± 1.91% on a dataset of 348 radiology reports (average bag-of-words similarity). In terms of clinical variable extraction (11 variables), the proprietary model ‘GPT-4o’ achieved an accuracy of 90% on a preliminary test dataset of 72 reports, while ‘GPT-3.5’ achieved 79%. Notably, the local, open-source models ‘Llama 3 70B’ and ‘Llama 3 8B’ achieved accuracies of 85% and 84%, respectively. The processing time per document was approximately six seconds. Certain variables were extractable at significantly higher average accuracies, e.g., affected side (97–100%), presence of joint effusion (92–99%), or anterior cruciate ligament pathology (90–94%).
Discussion and conclusions: The proposed pipeline enables efficient, fully automated digitization of medical documents while maintaining high extraction accuracy. Local, open-source LLMs achieved performance approaching leading proprietary models. This has significant implications for medical research and care delivery by making previously inaccessible datasets usable—such as transforming existing records into structured registries.




