70. Jahrestagung der Deutschen Gesellschaft für Medizinische Informatik, Biometrie und Epidemiologie e.V.
70. Jahrestagung der Deutschen Gesellschaft für Medizinische Informatik, Biometrie und Epidemiologie e.V.
Evaluating the Utility of Synthetic Data in Medical Machine Learning Using Pairwise Correlation Distance
Text
Introduction: The development of predictive models for medical applications is often hindered by the scarcity of high-quality datasets, particularly for rare diseases with limited documented cases. Incorporating synthetic data (SD) generated by advanced generative models (GMs) presents a promising solution to this challenge, however, their adoption in the medical field remains limited due to concerns about data fidelity and practical utility [1]. This study aims to address these uncertainties by rigorously evaluating the potential of SD for medical ML. Specifically, we investigate whether Pairwise Correlation Distance (PCD) can serve as a reliable and practical utility metric for assessing the quality and predictive value of SD in medical ML applications. PCD quantifies the similarity between synthetic and real datasets by measuring the difference in their pairwise feature correlation matrices [2].
Methods: The study utilizes the PIMA Indians Diabetes database [3], compromising eight laboratory features for 768 female patients, along with a binary label. We implemented three GMs and tuned their hyperparameters, using statistical similarity measures: CopulaGAN, Conditional-Tabular-GAN (CTGAN), and Tabular Variational Autoencoder (TVAE) [4], [5]. SD quality and utility are evaluated through a comprehensive correlation analysis based on two complementary approaches: (1) statistical similarity measures, and (2) classifier-based assessment, where models are trained on SD and tested on real data (RD) to evaluate predictive performance. Empirically, we derive a statistical metric indicative of SD utility and suitable for optimizing GM hyperparameters. Further extensive and safeguarded evaluation of SD realism aims to support the rationale for the SD application in medical ML.
Results: Table 1 [Tab. 1] presents intermediate results that illustrate the general trend observed. The hyperparameters of the GMs are optimized based on the achieved PCD to guide model selection. The SD generated by the optimized GMs is then used to train classifiers, subsequently evaluated on test RD using AUROC. Classifier performance when trained on RD is reported as the baseline. We report the percentage difference in AUROC relative to the baseline (∆ baseline), to default GM configuration (∆ default), and the PCD of optimized and default GMs (PCD[default, optimized]).
Table 1: Classification performance (AUROC) trained on SD by PCD-optimized GMs, evaluated on RD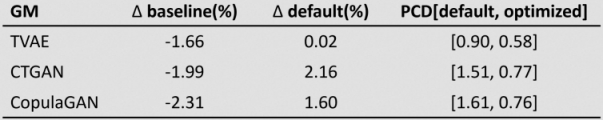
Particularly both evaluated GANs benefit from PCD-based parameter optimization. While the presented experiment employs SD and RD of equal size, evaluating tenfold SD size consistently shows congruent classifier performance to the baseline.
Conclusion: Our findings demonstrate that the PCD serves as a reliable and computationally efficient utility metric for SD across all evaluated generative models, as evidenced by strong correlation with classification performance. Furthermore, using PCD-based GM optimization provides classification results that are competitive with, or even superior to, those achieved using RD. However, it is important to note that PCD primarily captures linear correlations and may be insensitive to domain-specific or nonlinear dependencies. Despite this limitation, the consistently strong classification performance and high-quality SD produced using PCD support the viability of this approach. Although beyond the scope of present study, preliminary analysis suggests that PCD may contribute to a future framework for balancing privacy and utility in SD generation.
The authors declare that they have no competing interests.
The authors declare that an ethics committee vote is not required.
Literatur
[1] Kaabachi B, Despraz J, Meurers T, Otte K, Halilovic M, Kulynych B, Prasser F, Raisaro JL. A scoping review of privacy and utility metrics in medical synthetic data. NPJ Digit Med. 2025 Jan 27;8(1):60. DOI: 10.1038/s41746-024-01359-3[2] Goncalves A, Ray P, Soper B, Stevens J, Coyle L, Sales AP. Generation and evaluation of synthetic patient data. BMC Med Res Methodol. 2020 May 7;20(1):108. DOI: 10.1186/s12874-020-00977-1
[3] National Institute of Diabetes and Digestive and Kidney Diseases. Pima Indians Diabetes Database. [Accessed 2025 Apr 25]. Available from: https://www.kaggle.com/datasets/uciml/pima-indians-diabetes-database
[4] Patki N, Wedge R, Veeramachaneni K. The Synthetic Data Vault. In: 2016 IEEE International Conference on Data Science and Advanced Analytics (DSAA); 2016 Oct 17-19; Montreal, QC, Canada. p. 399-410. DOI: 10.1109/DSAA.2016.49
[5] Xu L, Skoularidou M, Cuesta-Infante A, Veeramachaneni K. Modeling Tabular Data Using Conditional GAN [Preprint]. arXiv. 2019. DOI: 10.48550/arXiv.1907.00503




