70. Jahrestagung der Deutschen Gesellschaft für Medizinische Informatik, Biometrie und Epidemiologie e.V.
70. Jahrestagung der Deutschen Gesellschaft für Medizinische Informatik, Biometrie und Epidemiologie e.V.
Are Today’s LLMs Ready to Take Over SNOMED Coding?
Text
Introduction: High-quality semantic mapping to standardized medical terminologies like SNOMED is critical for interoperability and secondary use of health data. Traditionally, this task is performed manually by experienced coders. However, the recent advances in large language models (LLMs) have sparked interest in automating this process [1]. This study investigates whether LLMs can approximate or even match expert-level terminology coding quality, using the German Corona Consensus Dataset (GECCO) as a benchmark [2].
Methods: We selected a subset of 23 medical elements from the GECCO dataset. LLMs should provide the SNOMED Fully Specified Name (FSN) along with the corresponding code. Three established prompting strategies were tested: few-shot examples, chain-of-thought (CoT) prompting, and prompts designed to explicit reasoning [3]. Each strategy was applied to LLMs widely used in the medical domain: Claude 3.5 Haiku (Anthropic) and ChatGPT-4o (OpenAI), both accessed via their respective web-based platforms, as well as Llama 3.1 8b (Meta), and Mixtral 8x7b instruct v0.1 (Mistral AI SAS), which were run locally using LM Studio (macOS, Apple M3 Pro, March 2025 version). Model outputs were assessed for correctness by comparing both the FSN and the SNOMED code to the GECCO reference, using the ISO/TS 21564 Equivalence Assessment Score [4]. The ISO scores were then benchmarked against mappings provided by two experienced human coders, which served as a high-confidence gold standard [5]. The top five outputs were statistically compared against the lowest-performing model that was still eligible for inclusion in the analysis, using the Mann–Whitney U test. Results with a p-value below 0.05 were considered statistically significant.
Results: As to be seen in Figure 1 [Fig. 1], none of the tested LLMs matched the performance of the human-generated gold standard. The gold standard achieved a mean ISO score of 0.68, with SNOMED codes predominantly categorized as equivalent to the medical element (n = 13). The best-performing LLM (GPT-4o, using a reasoning-based prompt) achieved a mean ISO score of 2.95, with codes classified as equivalent in only five instances. Of the evaluated elements 31% contained both an existing and correctly matching FSN and code. All models produced a considerable number of hallucinated outputs that did not exist. None of the locally run models contained a single pair of existing and matching FSN and code. Among the statistical comparisons, only the gold standard differed significantly from the lowest-performing model (p-value < 0.01).
Figure 1: Top five outputs by mean ISO Score; NaN values displayed as ISO Score: 4; Mean (black); Median (grey); existing SNOMED codes (black points), halluzinated SNOMED codes (red points)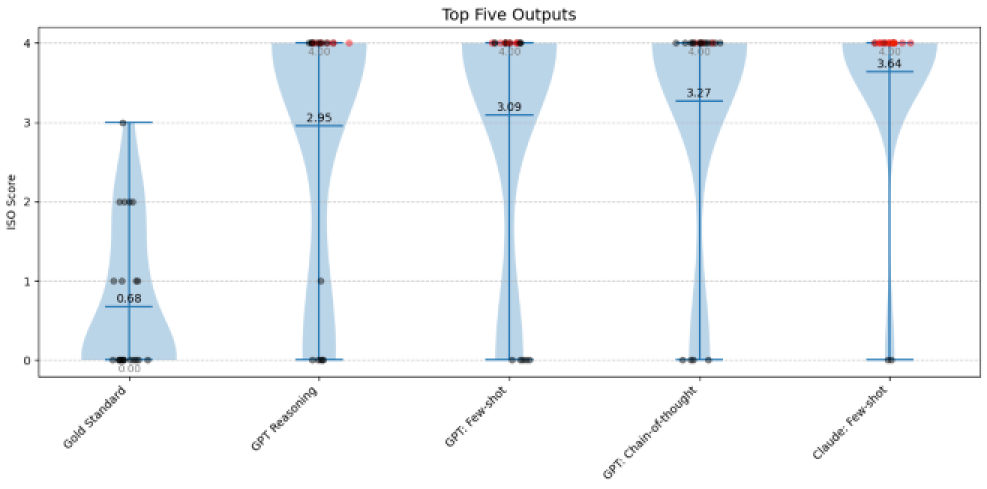
Conclusion: LLMs offer promising support across a wide range of medical applications. However, their tendency to hallucinate and lack of precision currently make them unreliable for SNOMED coding tasks. Accurate coding with SNOMED demands domain expertise and meticulous validation – capabilities LLMs have yet to match. Domain-specific training could lead to substantial improvements, but ongoing benchmarking against curated gold standards remains essential to assess the future role of AI in clinical coding support.
The authors declare that they have no competing interests.
The authors declare that an ethics committee vote is not required.
References
[1] Mustafa A, Naseem U, Rahimi Azghadi M. Large language models vs human for classifying clinical documents. Int J Med Inform. 2025 Mar;195:105800. DOI: 10.1016/j.ijmedinf.2025.105800[2] Sass J, Bartschke A, Lehne M, Essenwanger A, Rinaldi E, Rudolph S, Heitmann KU, Vehreschild JJ, von Kalle C, Thun S. The German Corona Consensus Dataset (GECCO): a standardized dataset for COVID-19 research in university medicine and beyond. BMC Med Inform Decis Mak. 2020 Dec 21;20(1):341. DOI: 10.1186/s12911-020-01374-w
[3] Schulhoff S, et al. The Prompt Report: A Systematic Survey of Prompt Engineering Techniques [Preprint]. arXiv. 2025 Feb 26. DOI: 10.48550/arXiv.2406.06608
[4] International Organization for Standardization. ISO/TS 21564. Health Informatics – Terminology resource map quality measures (MapQual). [Accessed 2025 Apr 25]. Available from: https://www.iso.org/standard/71088.html
[5] Vorisek CN, Klopfenstein SAI, Sass J, Lehne M, Schmidt CO, Thun S. Evaluating Suitability of SNOMED CT in Structured Searches for COVID-19 Studies. Stud Health Technol Inform. 2021 May 27;281:88-92. DOI: 10.3233/SHTI210126




