German Congress of Orthopaedics and Traumatology (DKOU 2025)
Deutscher Kongress für Orthopädie und Unfallchirurgie 2025 (DKOU 2025)
Bioinformatics and machine learning-based prognostic modeling and immune infiltration revealing key genes associated with Li-Fraumeni syndrome and osteosarcoma
Text
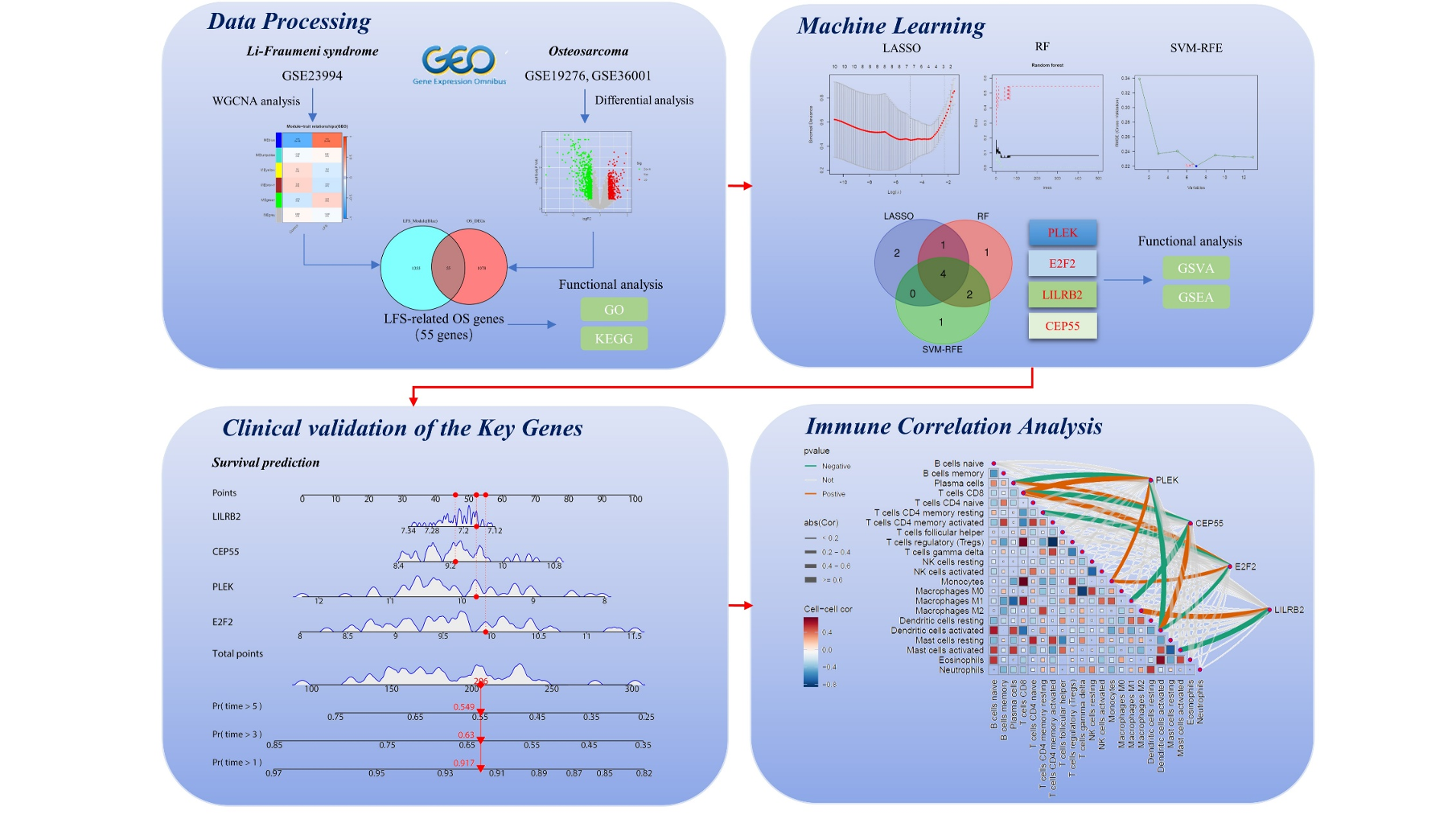
Objectives and questions: Li-Fraumeni syndrome (LFS) is a cancer susceptibility syndrome in which osteosarcoma (OS) accounts for the third highest incidence of LFS malignancies. This study aims to identify the key genes associated with the susceptibility of LFS patients to OS, establish a prognostic model, and investigate the role of immune cells in this process. It provides theoretical support for subsequent clinical treatment and prognostic analysis.
Material and methods: We downloaded the microarray datasets of LFS (GSE23994) and OS (GSE19276, GSE36001, GSE21257) from the Gene Expression Omnibus (GEO) database. Differential expression genes (DEGs) analysis and weighted gene co-expression network analysis (WGCNA) were applied to identify LFS-related genes in OS. Functional enrichment analysis was used to reveal the underlying mechanisms. Then, key genes were identify using protein-protein interactions (PPIs) and three machine learning algorithms, the Least Absolute Shrinkage Selection Operator (LASSO), Support Vector Machine-Recursive Feature Elimination (SVM-RFE), and Random Forest (RF). The diagnostic value of the model was assessed by plotting receiver operating characteristic (ROC) curves and nomogram, followed by prognostic analysis. Finally, single-gene gene set variation analysis (GSVA), gene set enrichment analysis (GSEA), immune cell infiltration analysis, and correlation analysis were conducted for key genes.
Results: A total of 55 genes were identified as LFS-related genes in OS. Functional analysis indicated that the potential pathogenic mechanisms of these genes were highly associated with cell cycle regulation and the AMPK signaling pathway. Furthermore, four key genes (PLEK, CEP55, E2F2, and LILRB2) were identified using three machine learning algorithms. A diagnostic model (AUC > 0.9) and a survival prognostic model were established based on these four key genes for identifying OS in LFS patients. Finally, immune infiltration analysis, along with single-gene GSEA and GSVA analyses, suggested that these genes might be associated with the dysregulation of immune cells, particularly M0 macrophages, and could regulate OS development through multiple pathways, including involvement in the cell cycle and DNA replication.
Discussion and conclusions: We identified PLEK, CEP55, E2F2, and LILRB2 as key biological genes in the occurrence and progression of OS in LFS patients using bioinformatics and machine learning algorithms. Based on these findings, we successfully constructed a novel diagnostic and survival prediction model and analyzed the potential regulatory mechanisms of these key genes. Our findings provide some support for further clinical studies and provide new perspectives for subsequent clinicians to treat patients with LFS-related OS.
Figure 1 [Fig. 1]