German Congress of Orthopaedics and Traumatology (DKOU 2025)
Deutscher Kongress für Orthopädie und Unfallchirurgie 2025 (DKOU 2025)
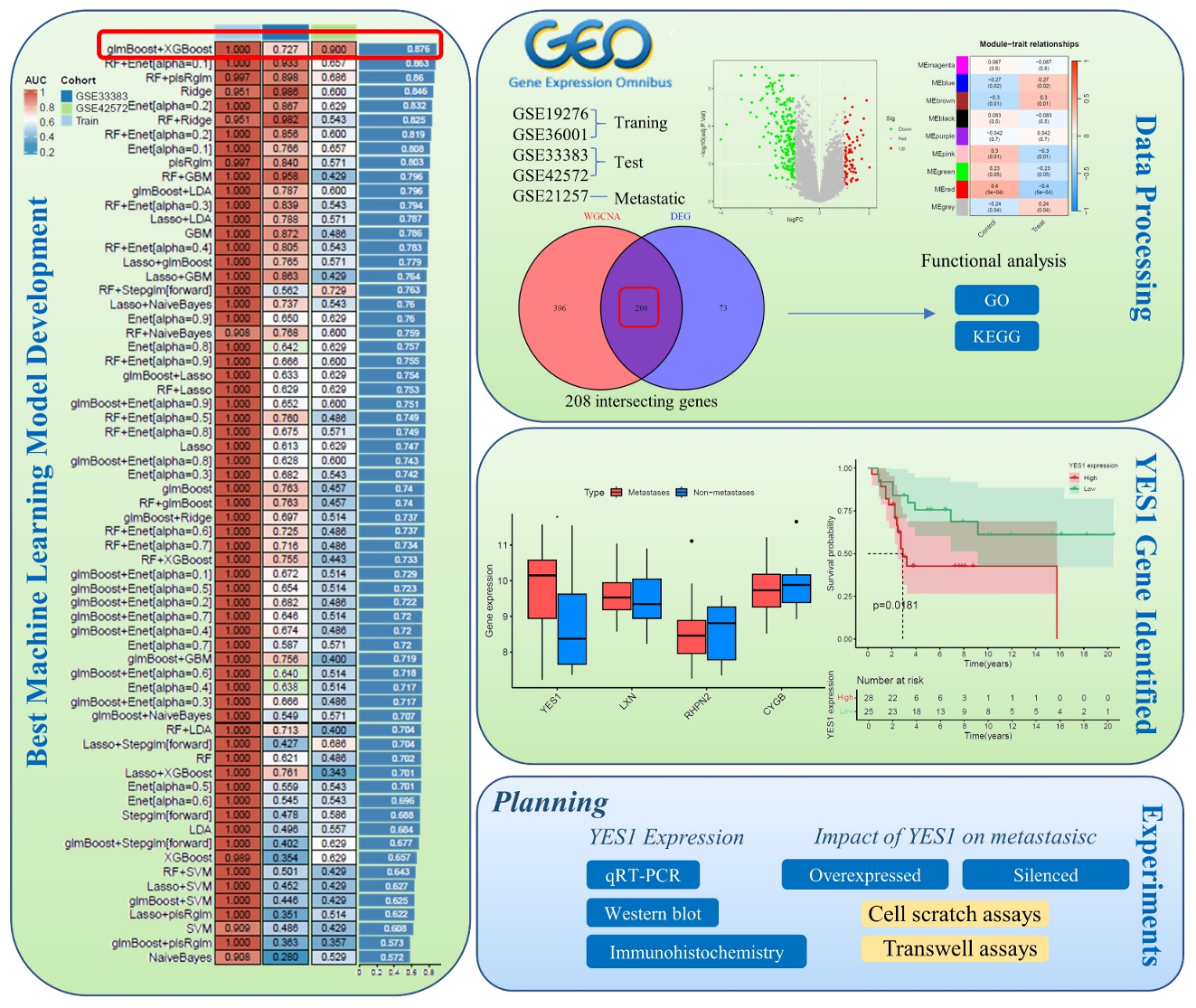
Revealing the role of the YES1 gene in regulating steosarcoma occurrence and metastasis based on bioinformatics and machine learning
Text
Objectives and questions: Osteosarcoma (OS) is a common primary malignant bone tumor in adolescents that is highly aggressive and metastatic. The YES1 gene has been shown to be associated with metastasis of various cancers, but its role in OS remains unclear. This study aimed to elucidate the mechanism of YES1 in OS metastasis by cellular experiments and machine learning algorithms.
Material and methods: In this study, 113 machine learning models were used to investigate the molecular basis of YES1 to promote OS transfer. We obtained OS-related datasets (GSE19276, GSE36001, GSE33383, GSE42572, and GSE21257) from the GEO database and identified OS-related differentially expressed genes (DEGs). Weighted Gene Co-expression Network Analysis (WGCNA) was used to determine OS-related gene modules. Machine learning models were applied to construct the optimal predictive model, highlighting key genes. The potential role of YES1 and its associated genes in OS metastasis was evaluated through Kaplan-Meier (KM) survival analysis, Gene Set Enrichment Analysis (GSEA), and immune infiltration analysis. Next, we will analyze the expression of YES1 and related pathways in U2OS cell lines using qRT-PCR, Western blotting, and immunohistochemistry (IHC). At the same time, the effects of normal, overexpressed, and silenced YES1 on OS cell migration and invasion will be evaluated using cell scratch tests, transwell tests, and flow cytometry.
Results: The study identified 281 DEGs in the test group OS dataset. WGCNA revealed that the most relevant red module for OS contained 604 key genes. Among the 113 machine learning models tested, the glmBoost + XGBoost combination obtained the highest average area under the curve (AUC) of 0.876 in both the training and test groups and selected 10 key genes. Receiver operating characteristic curve (ROC) analysis showed that CYGB, LXN, RHPN2 and YES1 had strong disease prediction ability (AUC > 0.7). Meanwhile, YES1 gene was highly expressed in the OS metastasis group (p<0.05), and related gene, GSEA and immune infiltration analyses indicated that YES1 might promote OS metastasis through the cell cycle, T-cell CD4 memory quiescence, and γδ T-cell involvement. As YES1 can promote cancer cell metastasis through the PI3K/AKT pathway in other tumors, we speculated that it might be similar in OS.
Discussion and conclusions: The results showed that patients with high YES1 expression had shorter survival time and YES1 may promote OS metastasis by regulating the PI3K/AKT pathway. These findings provide new insights and support for clinical treatment of OS metastases.
Figure 1 [Abb. 1]